Visualisation of spatial resources with TemplateFlow¶
This notebook showcases how TemplateFlow can facilitate visualisation of spatially standardised data, including template and atlas resources. We begin by importing the TemplateFlow API, along with nibabel for reading neuroimaging data files, matplotlib for plotting, and a number of plotting functions from the nilearn and niworkflows packages.
[1]:
import templateflow.api as tflow
import nibabel as nb
import matplotlib.pyplot as plt
from niworkflows.viz.utils import (
plot_registration,
plot_segs,
compose_view,
cuts_from_bbox
)
from IPython.display import SVG, display
/usr/local/anaconda3/lib/python3.7/site-packages/sklearn/linear_model/least_angle.py:30: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
method='lar', copy_X=True, eps=np.finfo(np.float).eps,
/usr/local/anaconda3/lib/python3.7/site-packages/sklearn/linear_model/least_angle.py:167: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
method='lar', copy_X=True, eps=np.finfo(np.float).eps,
/usr/local/anaconda3/lib/python3.7/site-packages/sklearn/linear_model/least_angle.py:284: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
eps=np.finfo(np.float).eps, copy_Gram=True, verbose=0,
/usr/local/anaconda3/lib/python3.7/site-packages/sklearn/linear_model/least_angle.py:862: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
eps=np.finfo(np.float).eps, copy_X=True, fit_path=True,
/usr/local/anaconda3/lib/python3.7/site-packages/sklearn/linear_model/least_angle.py:1101: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
eps=np.finfo(np.float).eps, copy_X=True, fit_path=True,
/usr/local/anaconda3/lib/python3.7/site-packages/sklearn/linear_model/least_angle.py:1127: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
eps=np.finfo(np.float).eps, positive=False):
/usr/local/anaconda3/lib/python3.7/site-packages/sklearn/linear_model/least_angle.py:1362: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
max_n_alphas=1000, n_jobs=None, eps=np.finfo(np.float).eps,
/usr/local/anaconda3/lib/python3.7/site-packages/sklearn/linear_model/least_angle.py:1602: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
max_n_alphas=1000, n_jobs=None, eps=np.finfo(np.float).eps,
/usr/local/anaconda3/lib/python3.7/site-packages/sklearn/linear_model/least_angle.py:1738: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
eps=np.finfo(np.float).eps, copy_X=True, positive=False):
/usr/local/anaconda3/lib/python3.7/site-packages/sklearn/decomposition/online_lda.py:29: DeprecationWarning: `np.float` is a deprecated alias for the builtin `float`. To silence this warning, use `float` by itself. Doing this will not modify any behavior and is safe. If you specifically wanted the numpy scalar type, use `np.float64` here.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
EPS = np.finfo(np.float).eps
/usr/local/anaconda3/lib/python3.7/site-packages/sklearn/feature_extraction/image.py:167: DeprecationWarning: `np.int` is a deprecated alias for the builtin `int`. To silence this warning, use `int` by itself. Doing this will not modify any behavior and is safe. When replacing `np.int`, you may wish to use e.g. `np.int64` or `np.int32` to specify the precision. If you wish to review your current use, check the release note link for additional information.
Deprecated in NumPy 1.20; for more details and guidance: https://numpy.org/devdocs/release/1.20.0-notes.html#deprecations
dtype=np.int):
/usr/local/anaconda3/lib/python3.7/site-packages/nilearn/datasets/__init__.py:89: FutureWarning: Fetchers from the nilearn.datasets module will be updated in version 0.9 to return python strings instead of bytes and Pandas dataframes instead of Numpy arrays.
"Numpy arrays.", FutureWarning)
Visualising a template¶
In addition to numerous human template resources, TemplateFlow includes the Fischer344 rodent template for use with preclinical imaging workflows. Let’s start by creating a mosaic plot of this template. To do this, we begin by using the TemplateFlow client to retrieve the T2-weighted image (suffix='T2w') of the Fischer template. We also retrieve a brain mask (desc='brain', suffix='mask'); this will be used to determine the sections of the brain to be visualised.
Note: Some templates might have several different resolutions; in this case, a resolution argument should be used to specify the desired resolution. (Resolutions are defined in the template_description.json metadata file.)
[2]:
uid = 'Fischer344'
nii = nb.load(tflow.get(uid, desc=None, suffix='T2w'))
mask_nii = nb.load(tflow.get(uid, hemi=None, desc='brain', suffix='mask'))
Next, we use the plot_registration function from niworkflows to create a mosaic SVG plot of the Fischer344 brain. This plot will include axial, sagittal, and coronal slices. The number of slices is set by n_cuts, and the positions of the slices is determined by the niworkflows utility function cuts_from_bbox.
[3]:
n_cuts = 7
cuts = cuts_from_bbox(mask_nii, cuts=n_cuts)
figs = plot_registration(
nii,
'fixed-image',
estimate_brightness=True,
cuts=cuts,
label=uid,
contour=mask_nii,
compress='auto',
dismiss_affine=False,
)
[4]:
for i, fig in enumerate(figs):
fig.save(f'/tmp/tpl-{uid}_view{i}.svg')
display(SVG(filename=f'/tmp/tpl-{uid}_view{i}.svg'))
Segmentation overlay¶
Most templates in TemplateFlow are distributed with segmentations. Discrete segmentations (suffix='dseg') are voxelwise annotations that assign each voxel of the brain to a single region or tissue class. Probabilistic segmentations (suffix='probseg') instead assign each voxel to a categorical distribution over tissue classes or regions.
Let’s visualise the tissue classes present in the Fischer344 brain. To do this, we’ll use a 4-way probabilistic segmentation into grey matter, white matter, cerebrospinal fluid, and a catch-all ‘other’ category to delineate boundary contours of each tissue class. We begin by retrieving the probabilistic segmentations using the TemplateFlow API. Now, we use another plotting function from niworkflows designed for this purpose, plot_segs, again selecting the slices to be plotted with
guidance from the mask (bbox_nii argument).
[5]:
probseg_nii = [str(path) for path in tflow.get('Fischer344', suffix='probseg')]
figs = plot_segs(
image_nii=str(tflow.get(uid, desc=None, suffix='T2w')),
seg_niis=probseg_nii,
bbox_nii=str(tflow.get(uid, hemi=None, desc='brain', suffix='mask')),
out_file=f'/tmp/tpl-{uid}_seg.svg'
)
[6]:
for i, fig in enumerate(figs):
fig.save(f'/tmp/tpl-{uid}_seg{i}.svg')
display(SVG(filename=f'/tmp/tpl-{uid}_seg{i}.svg'))
Atlas overlay¶
In addition to tissue-class annotations, many templates are distributed with atlases, which associate with each spatial coordinate a particular annotation or set of annotations. Atlases include inter alia anatomical parcellations (e.g., annotations of gyri and sulci) and functional parcellations (e.g., annotations of brain networks, or regions displaying homogeneous activity or preferential connectivity).
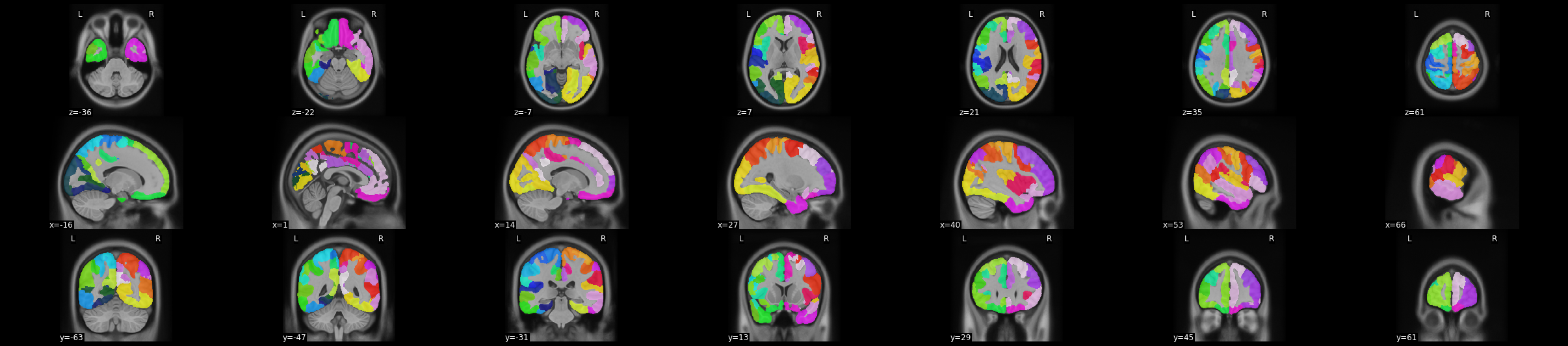
Here, we’ll visualise a functional atlas: the 100-region cortical parcellation of Schaefer and colleagues (2018), which is situated in the stereotaxic coordinate space of the MNI152NLin2009cAsym template. We use the atlas argument to the TemplateFlow API to specify the atlas family, and here we additionally use the desc argument to disambiguate among atlases within the family. (The Schaefer parcellation has several different resolutions and parcel numbering schemes, so this
disambiguation is required.) Finally, because the atlas deterministically assigns each voxel coordinate to a single parcel, we again specify suffix='dseg'.
[7]:
from nilearn import plotting
uid = 'MNI152NLin2009cAsym'
nii = nb.load(tflow.get(uid, resolution=1, desc=None, suffix='T1w'))
atlas_nii = nb.load(tflow.get(
uid, resolution=1,
desc='100Parcels7Networks',
atlas='Schaefer2018',
suffix='dseg'
))
Now, we’ll use nilearn’s plot_roi function to create a mosaic view of the atlas. This function is used for visualising regions of interest (ROIs). We set the background image to be the template T1 we retrieved above and the ROI image to be the atlas.
[8]:
%matplotlib inline
plotting.plot_roi(roi_img=atlas_nii, bg_img=nii, display_mode='mosaic')
[8]:
<nilearn.plotting.displays.MosaicSlicer at 0x130e03c90>
Template-to-template transformations¶
To support the flow of spatial knowledge and annotations among template spaces, TemplateFlow aims to compute template-to-template transformations. These transformations would map each coordinate in a particular template’s coordinate space to its analogue in the spaces engendered by all other suitable templates. Such transformations could, for instance, be used to move the Schaefer parcellation above into the coordinate space of a pediatric or infant template, thereby enabling its use with additional study populations.
Here, we’ll visualise the product of such a template-to-template transformation. This transformation links the version of the MNI template distributed with FSL, MNI152NLin6Asym, to the ICBM’s MNI152NLin2009cAsym template. To visualise this registration, we’ll first use the template-to-template transform to warp the FSL template (MNI152NLin6Asym) into ICBM space (MNI152NLin2009cAsym). For this, we use the command-line tool antsApplyTransforms. We can use inline calls to the
TemplateFlow API to retrieve the template-to-template transform, the input template, and the reference template, and then use these to populate the call to antsApplyTransforms.
[9]:
%%bash
template_src='MNI152NLin6Asym'
template_dst='MNI152NLin2009cAsym'
transform=$(
python -c "
import templateflow.api as tflow
import re
xfm = [
path for path in tflow.get('${template_dst}', mode='image', suffix='xfm')
if re.search('.*from-${template_src}.*', str(path))
]
print(xfm[0])
"
)
input=$(
python -c "
import templateflow.api as tflow
print(tflow.get('${template_src}', desc=None, resolution=1, suffix='T1w'))
"
)
reference=$(
python -c "
import templateflow.api as tflow
print(tflow.get('${template_dst}', desc=None, resolution=1, suffix='T1w'))
"
)
output=/tmp/tpl-${template_src}_space-${template_dst}_res-01_T1w.nii.gz
echo ${ANTSPATH}/antsApplyTransforms \
-d 3 -e 0 \
-i ${input} \
-r ${reference} \
-o ${output} \
-n LanczosWindowedSinc \
-t ${transform}
/antsApplyTransforms -d 3 -e 0 -i /Users/rastko/.cache/templateflow/tpl-MNI152NLin6Asym/tpl-MNI152NLin6Asym_res-01_T1w.nii.gz -r /Users/rastko/.cache/templateflow/tpl-MNI152NLin2009cAsym/tpl-MNI152NLin2009cAsym_res-01_T1w.nii.gz -o /tmp/tpl-MNI152NLin6Asym_space-MNI152NLin2009cAsym_res-01_T1w.nii.gz -n LanczosWindowedSinc -t /Users/rastko/.cache/templateflow/tpl-MNI152NLin2009cAsym/tpl-MNI152NLin2009cAsym_from-MNI152NLin6Asym_mode-image_xfm.h5
(We’ve conveniently placed the output of this transformation in the data subdirectory.)
Next, we’ll overlay the warped MNI152NLin6Asym template (the moving image) and the MNI152NLin2009cAsym template (the fixed image or target of transformation) to facilitate assessment of the quality of the transformation. Once again, we can use the client to obtain the fixed image and its corresponding mask.
[10]:
fixed_uid = 'MNI152NLin2009cAsym'
moving_uid = 'MNI152NLin6Asym'
fixed_nii = nb.load(tflow.get(fixed_uid, resolution=1, desc=None, suffix='T1w'))
moving_nii = nb.load('data/tpl-MNI152NLin6Asym_space-MNI152NLin2009cAsym_res-01_T1w.nii.gz')
mask_nii = nb.load(tflow.get(fixed_uid, resolution=1, desc='brain', suffix='mask'))
As above, we use the plot_registration utility from niworkflows, calling the utility on both the fixed and the moving image. This time, we pass both plots to the compose_view function from niworkflows,
[11]:
n_cuts = 7
cuts = cuts_from_bbox(mask_nii, cuts=n_cuts)
reg_fig = compose_view(
plot_registration(
fixed_nii,
'fixed-image',
estimate_brightness=True,
cuts=cuts,
label=fixed_uid,
contour=mask_nii,
compress='auto',
dismiss_affine=False,
),
plot_registration(
moving_nii,
'moving-image',
estimate_brightness=True,
cuts=cuts,
label=moving_uid,
contour=mask_nii,
compress='auto',
dismiss_affine=False,
)
)
[12]:
display(SVG(filename=reg_fig))